Discovering the functional genomics of adaptation
Half a century after the Green Revolution, we have learned that monumental yield increases in staple crops such as rice and wheat are attributed to specific mutations in a handful of genes. Later still we discovered that similar, naturally occurring mutations in wild plants can produce similar phenotypes. These discoveries demonstrate the power of a retroactive analysis to uncover the genes that changed history. Now, in the age of genomics and gene editing, we are poised to discover the genes that will define the future, by addressing the challenge of identifying genes responsible for adaptation at both genomic scales and functional molecular resolutions.
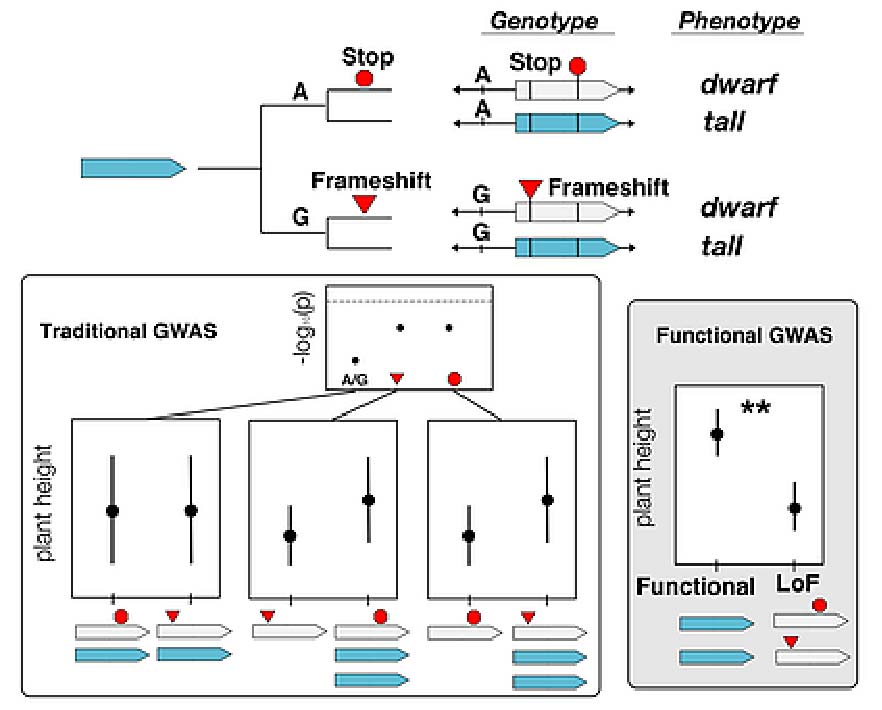
Traditional genome wide association scan (GWAS) approaches often fail to identify causative loci because of allelic heterogeneity and lack predictive power because of epistasis. Traditional GWAS is also intended to identify genome regions of interest rather than functionally definitive causative mutations, meaning that results cannot be readily translated to applied outcomes. Alternatively, a genome-wide association framework that contrasts alleles defined by functional state rather than linked SNPs can overcome allelic heterogeneity, provide a basis to empirically study epistasis, and identify targets immediately useful for hypothesis testing (Figure 1).
We have developed such a functional GWAS approach and used it to study drought adaptive loss-of-function alleles in Arabidopsis thaliana (Monroe et al. 2018, eLife). While in Arabidopsis, we have taken advantage of T-DNA knockout lines to study drought adaptive flowering time evolution, future work will extend this approach across plant species, traits, and functional allele classes and include the use of CRISPR based gene editing.
Novel approaches linking genes, environments, and traits
To understand the plant genomics of climate adaptation, it is important to study how genomes interact with the environment. For example, variation in functional genomic diversity along environmental gradients reflects the history of evolutionary processes such as natural selection. And during plant development, the genome provides the blueprint for phenotypic responses to environmental perturbation. By studying natural selection and development together, a more mechanistic understanding of climate adaptation can be developed that links genes, environments and traits.
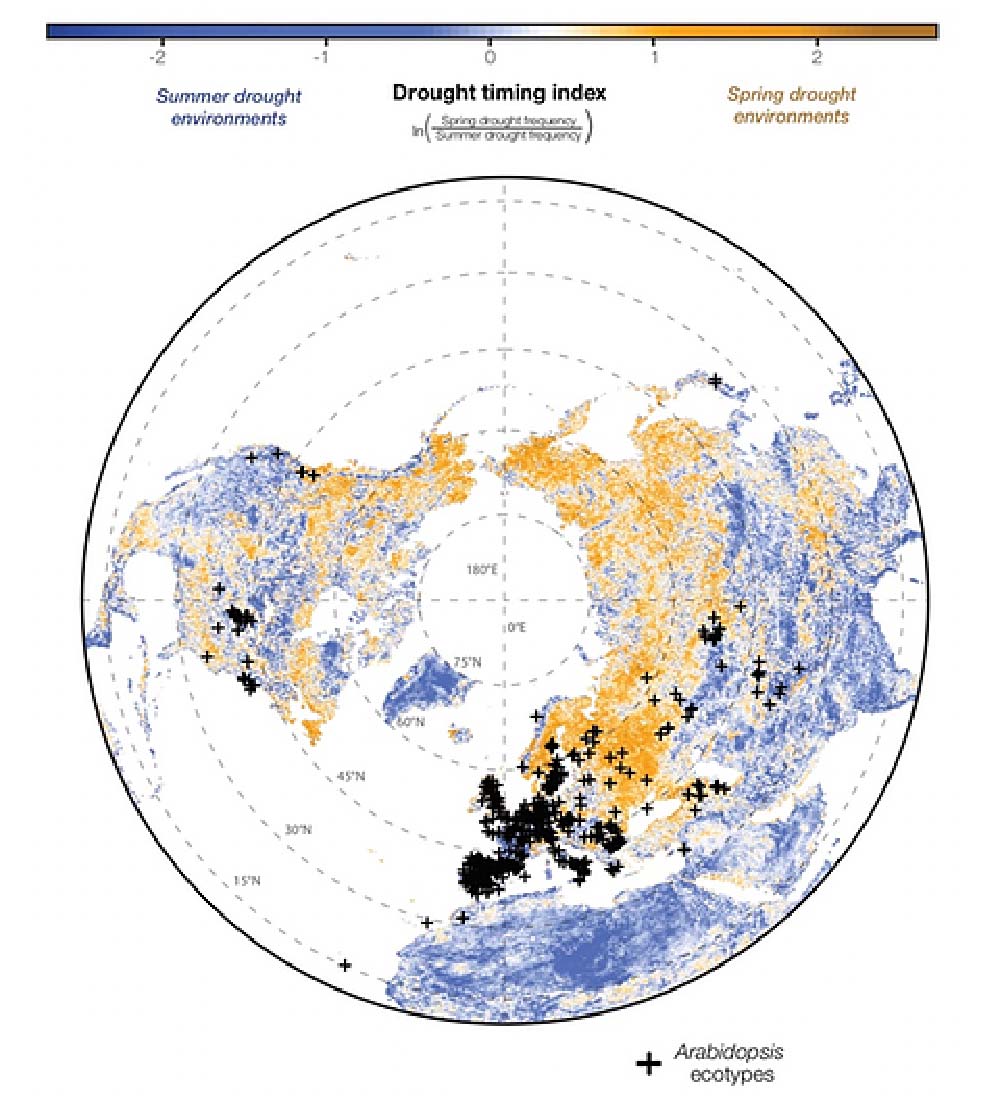
Locally adapted populations of wild plant species and land races of crops present opportunities to exploit allele-environment associations to identify genes responsible for climate adaptation. We have applied such a framework to study the genetic basis of adaptation to different dimensions of climate, such as winter temperatures in Arabidopsis (Monroe et al. 2016, Molecular Ecology) and found that allele-environment associations readily predict allele-phenotype associations (Monroe et al. 2018, eLife). This approach highlights the ability to use environmental gradients to find causal loci for traits that are particularly difficult or expensive to measure. It is critical to study ecologically relevant environmental gradients. To study the genomic and phenotypic basis of drought adaptation we have pioneered the use of satellite remote sensing to characterize historic drought regimes and applied this across a number of systems (Mojica et al. 2016, Plant Science; Ditttberner et al. 2018 Molecular Ecology; Monroe et al. 2018, eLife; Monroe et al. 2019, New Phytologist).
Historically, plant responses to environmental stress have been studied by growing plants under two contrasting treatments (i.e. “wet” and “dry”). While the simplicity of this approach is experimentally attractive, it overlooks the quantitative nature of plant environments and non-linear plant responses which provide necessary resolution to understand the mechanisms of stress tolerance. Working with Dave Des Marias at MIT, we are designing new experimental and statistical approaches to study plant responses as a function of a continuous gradient in water availability in the model grass Brachypodium.
Applying genomics to investigate the ecosystem consequences of climate adaptation
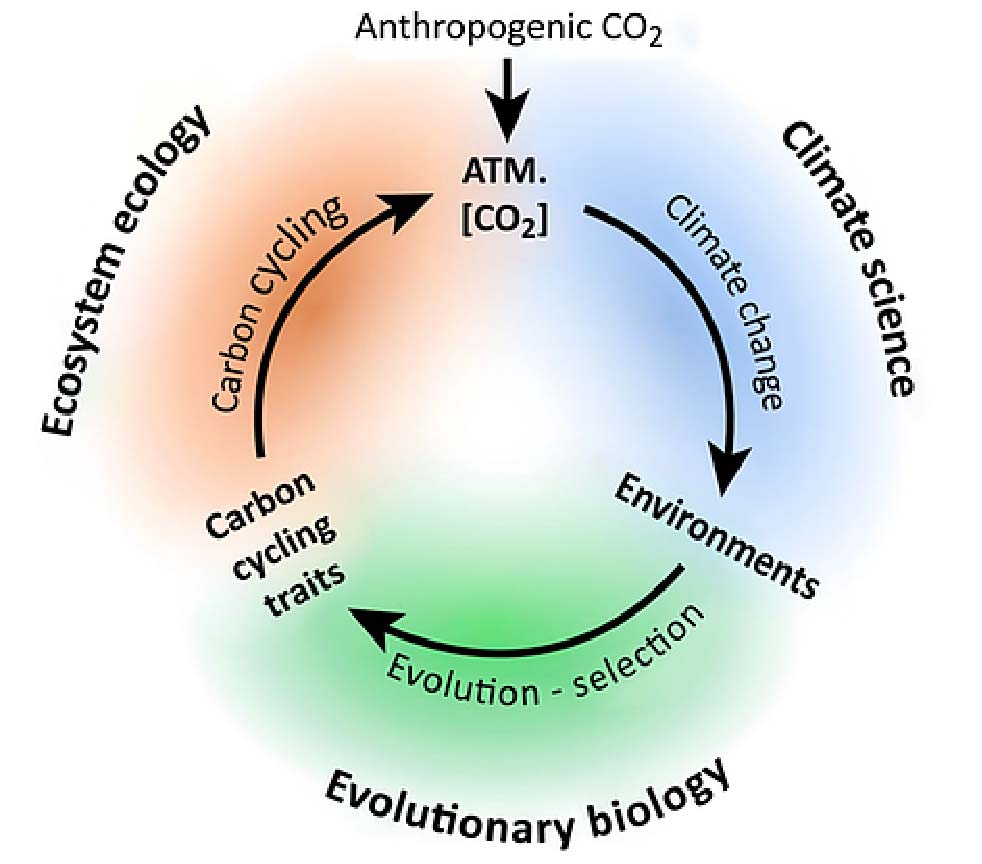
In 1881, twenty-two years after On the Origin of Species, Charles Darwin published a book inspired by the idea that organism adaptation can significantly impact the environment. Since then, mounting evidence indicates that adaptive evolution, especially in plants, has altered ecosystems and the climate throughout Earth’s history. Furthermore, recent work has indicated the possibility for rapid adaptation in response to anthropogenic climate change to significantly alter carbon cycling (Monroe et al. 2018 Trends in Ecology and Evolution). Studying this process is important for predicting future atmospheric carbon dioxide concentrations, developing agricultural systems that maximize ecosystem services, and is at the forefront of providing a much-needed synthesis between evolutionary and ecosystem ecology.
Our research has been supported by the NSF, USDA NIFA, ERC, HATCH, UC Davis, Colorado State University, and the Max Planck Society.